José Alonso wrote on mitk-users:Hello MITKers!
I am trying to package and distribute a .dmg file of my application for some users who work on macOS. The project is building successfully, and the make package too.
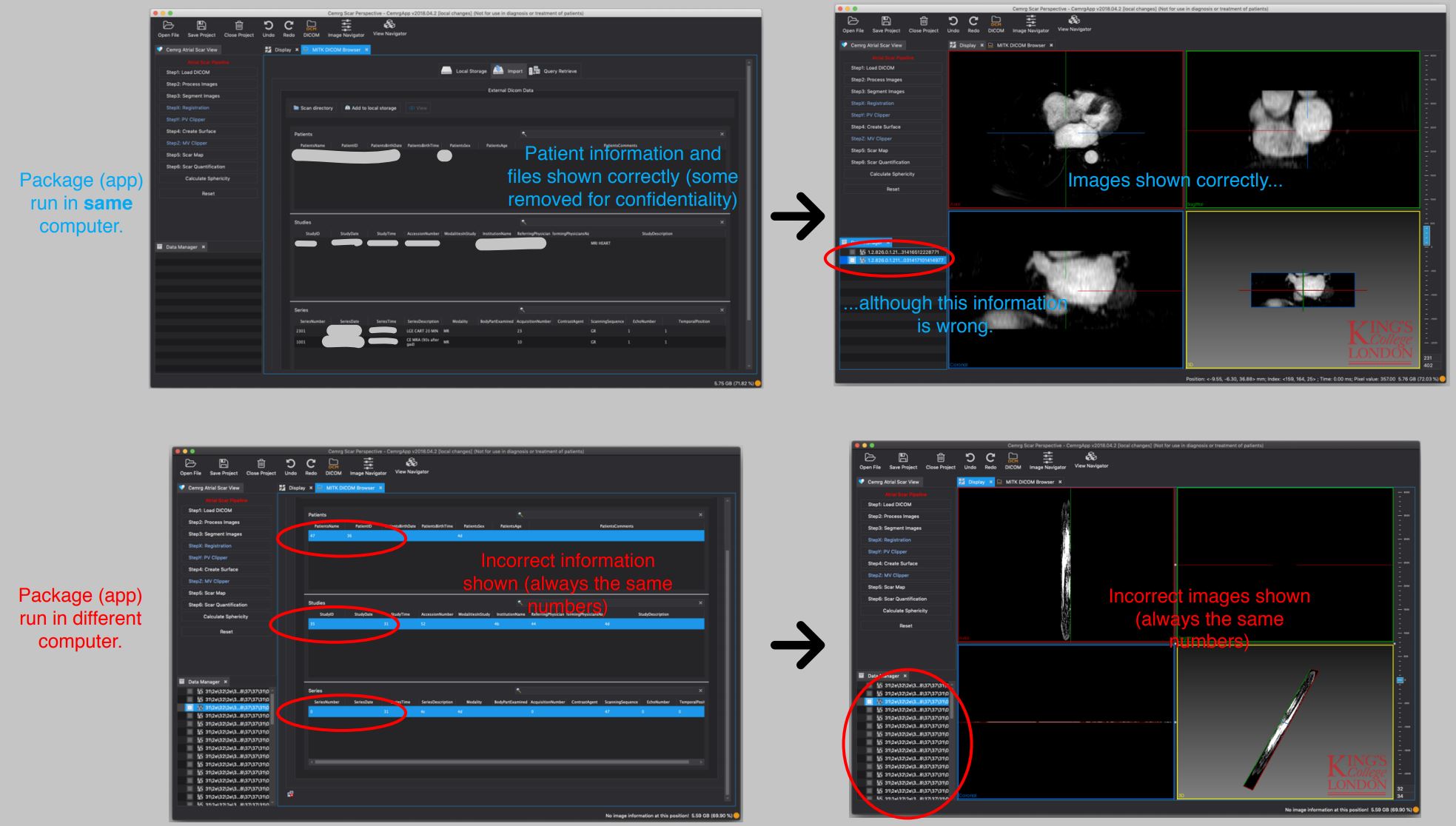
We are finding a problem when using the DICOM reader. In the computer I built the package, this works perfectly, however when I take the .dmg file and give it to one of our users, the DICOM reader shows incorrect information and does not load any of the relevant files.
How to reproduce the error:
- Open the DICOM reader.
- Click Scan directory.
I have tried the super build both with the Debug and Release options with no luck. Is there anything else we should need to do?
I am sharing screenshots I took of both cases in the attached file for you to see what we mean.
Description
Description
| Status | Assigned | Task | ||
|---|---|---|---|---|
| Resolved | kislinsk | T27196 Rewrite packaging of MITK | ||
| Resolved | kislinsk | T26451 DICOM plugin cannot parse DICOM directories in macOS installer | ||
| Resolved | kislinsk | T26164 DICOM Query/Retrieve doesn't work at least in Ubuntu release installers |
Event Timeline
Comment Actions
It would be helpfull to see the log output in order to see if a different reader configuration was choosen and which. Please activate the log window and check the out put of the read process.
There should be outputs like this for each checked configuration. The reader always picks the configuration with the lowest number of blocks or the first that offers one block.
Comment Actions
I tried using the Logging as @floca suggested, however I am not familiar with what I should be looking for (attached MitkWorkbench_.log file). I tried downloading the MITK Workbench binaries corresponding to v2018.04.2 and got the same error, attaching the screenshot.
Comment Actions
@jsolislemus Thanks for the info. If I understood it correctly also the official installer shows this problem. Could you please do me a favor and check with the official installer if you have the same loading problem, if you do not use the DICOM browser, but instead the normal file open dialog or dragNdrop. With this information we can narrow it down a bit more.
Thanks.
Comment Actions
Hello, sure. Please let me know if you need clarification, I am writing this with some haste, so I might not be as thorough or clear in my explanations.
(1) Using the Open File button -> Selected the (autoselect) -> NOT clicking on "apply the same to the remaining files" (file MitkWorkbench_MRIreadeAutoselect.log and looking as in the screenshot attached). The images were loaded individually, and not as a single dicom.
(2) Using the Open File button -> Selected the 3D reader -> NOT clicking on "apply the same to the remaining files" (got an error message, screenshot of error message attached)
Comment Actions
Thanks.
Regarding this workflow: I have another question. How many files did you select in the dialoge? One file of the stack as selection would be sufficient.
Comment Actions
Hm, ok. Then the log output is somehow inconsistent, with what is loaded into the workbench.
@kislinsk How do one enforces the more detailed log? The logging console is capable to show more information then given in the attached log.
Two further questions:
- Does the (a) self generated installer package and (b) the MITKWorkbench work on the system you use to generate (a) or does the installer (one or both) work not?
- Could you please test if the MITKWorkbench installer for other systems show the same problems.
I am looking for indication if it is a problem more related to a specific system setup or the package setup.
Thanks.
Comment Actions
Apologies for the delay in response..
Apologies, but I am not sure I understand your first questions, but given your last statement:
I am looking for indication if it is a problem more related to a specific system setup or the package setup.
The problem occurs (so far) when bringing a packaged version of the app (like the MITKWorkbench) from computer A, where the project was built, to computer B, where someone else wants to run the app. We have tested this on Ubuntu (18.04 and 16.04) and macOS (10.14).
That is, computer A is perfectly capable of running the packaged version. The problem comes only on computer B.
Comment Actions
No problem. A now fixed typo made it hard to understand.
As far as I have understood the Following is the case:
- On computer B your own packaged version does not work (message from 8/29)
- On computer B the downloaded binaries does not work (message from 7/31).
My question was, which of the versions (own, downloaded) work on computer A?
Comment Actions
Hello,
I got some more feedback from one of our users which I think might be related to this problem.
Commonly,DICOM images of respiratory navigated MRI sequences are stored with images of the respiratory navigator within the same folder. These are commonly called “localisers”. These extra files seen to be a problem when importing the DICOM and need to be removed manually in order for the sequence to be recognised.
Basically, some MRI sequences have other files stored in the same folder. In MITK v2016.03 these extra files were being ignored, however in v2018.04.2 these files cause a problem to the DICOM reader. I wonder if it is all related.
Comment Actions
Sorry for the long delay. Localisers should not be a problem per se. We also have such images in our protocols.
I must admit that only by "remote diagnistics" I am running out of ideas as we never had the problems (to my knowledge).
Would it be possible e.g. to give us a anonymized example data set? So that we can check and debug it. For what ever reason it seems that the systems read different values from the DCM tags or tread them differently and therefore load the data in other ways; e.g. splitting of a series in volumes.
Do the systems differ in there language/locale settings or something like this. This should be coverd by our code, but you never know.
Comment Actions
We came to the conclusion that we need example data to reproduce this issue on our site. Can we arrange a transfer (see above)?
Comment Actions
Sorry for the long delay. Thank you for your help.
We can send some data, I might be able to get some data to share with you, including: (a) DICOM to test on a packaged version of MITK, as described at the beginning of this thread, and (b) some of our data that's failing with localisers. I think I can get it to you within a week or two.
could you let us know how could we arrange that?
Comment Actions
That would be great. Thanks!
Transfer: The most pragmatic way is the dkfz FTP-Upload for externals. I will send you details early next week, when I am back in the office.
Comment Actions
@jsolislemus : Could you provide me your email adresse? Then I will send you the instructions how to transfer data.
Thanks
Comment Actions
Hi, just to let you know I uploaded the data to the ftp with the instructions sent. I sent the full information about the location of the file to Ralf. Could you please confirm you are able to access it? Many thanks.
Comment Actions
We are currently looking into it. In this process we realized that we are not able to reproduce the proplems with the current master.
Do you have the posibility, to also check a build based on the current master. It would be interesting if the problem also exists in the master for your setup or is also not reproducable for you.
Comment Actions
I will try to build with the current master. I will post here my findings.
You should be able to reproduce the error with the MITK Workbench download, or building on version v2018.04.2.
Comment Actions
OK, will look into it. If you could feedback insights regarding the master, it would be great. Thanks.
Ralf
Comment Actions
We discussed this in the MITK meeting today and identified two possible reasons.
- DCMTK dictionary is missing in the installer, we should investigate this on a Mac @kislinsk you have one to test/examine this?
- Dynamic loading could possibly load libraries from the build tree if the installer is run on the same machine @jsolislemus Could you try to debug the dynamic loading process? This should work by setting DYLD_PRINT_LIBRARIES in the shell before running the application, don't have a Mac available right now
Comment Actions
Thank you @nolden , for your input. Apologies for the delay, I was needing to use my machine and could not get around to do the full compilation of the code with the master branch as @floca mentioned before (below the comment, for context)
Regarding the last post on the two possibilities for this:
- Could this be tested if an external DCMTK is built and called into MITK into the MITK_EXTERNAL_DCMTK variable?
- I can surely try this. Could you please let me know how to do the debug of the dynamic loading process? I would really appreciate if you could elaborate on this. Is this a command I need to run in the terminal before opening MITK? Is the application I should be opening the MITK Workspace?
Thank you very much, stay safe.
Comment Actions
@jsolislemus Sorry for the long delay, I kind of missed your comment and related discussions happened in T27669
Are you still affected? For 2. , I will try to provide you with proper command soon, for 1. I think this would probably make things more complicated since we don't really have use cases for external DCMTK
Comment Actions
Added T26164 as subtask for now, as it is probably the same issue (missing DCMTK dictionary).
Comment Actions
Hi, from what I've been reading, it seems this issue might be fixed? I read on T26264 that it might be fixed with a CMake variables related to the DCMTK dictionaries; although I'm not sure how could I implement it myself, while waiting for the next release (we're still developing on top of 2018.04.2). Thanks a lot for the effort, this error seemed very difficult to pinpoint.
Comment Actions
@jsolislemus There's a workaround if you can't wait for the next release which would also verify if your issue is fixed:
- Download https://raw.githubusercontent.com/InsightSoftwareConsortium/DCMTK/master/dcmdata/data/dicom.dic
- Open a terminal
- Type export DCMDICTPATH=/path/to/dicom.dic
- Start the workbench from the terminal: /path/to/MitkWorkbench.app/Contents/MacOS/MitkWorkbench
Comment Actions
I close this task as it is very likely fixed in T26164: DICOM Query/Retrieve doesn't work at least in Ubuntu release installers. Please re-open if it is not the case for you by using the workaround or building the latest develop version . :-)